California housing regression
In this notebook we’ll use the ITEA_regressor to search for a good expression, that will be encapsulated inside the ITExpr_regressor class, and it will be used for the regression task of predicting California housing prices.
[1]:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn import datasets
from sklearn.model_selection import train_test_split
from IPython.display import display
from itea.regression import ITEA_regressor
from itea.inspection import *
import warnings
warnings.filterwarnings(action='ignore', module=r'itea')
The California Housing data set contains 8 features.
In this notebook, we’ll provide the transformation functions and their derivatives, instead of using the itea feature of extracting the derivatives using Jax.
Creating and fitting an ITEA_regressor
[2]:
housing_data = datasets.fetch_california_housing()
X, y = housing_data['data'], housing_data['target']
labels = housing_data['feature_names']
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.33, random_state=42)
tfuncs = {
'log' : np.log,
'sqrt.abs' : lambda x: np.sqrt(np.abs(x)),
'id' : lambda x: x,
'sin' : np.sin,
'cos' : np.cos,
'exp' : np.exp
}
tfuncs_dx = {
'log' : lambda x: 1/x,
'sqrt.abs' : lambda x: x/( 2*(np.abs(x)**(3/2)) ),
'id' : lambda x: np.ones_like(x),
'sin' : np.cos,
'cos' : lambda x: -np.sin(x),
'exp' : np.exp,
}
reg = ITEA_regressor(
gens = 50,
popsize = 50,
max_terms = 5,
expolim = (0, 2),
verbose = 10,
tfuncs = tfuncs,
tfuncs_dx = tfuncs_dx,
labels = labels,
random_state = 42,
simplify_method = 'simplify_by_coef'
).fit(X_train, y_train)
gen | smallest fitness | mean fitness | highest fitness | remaining time
----------------------------------------------------------------------------
0 | 0.879653 | 1.075671 | 1.153701 | 1min17sec
10 | 0.794826 | 0.828574 | 0.983679 | 2min7sec
20 | 0.788833 | 0.799124 | 0.850923 | 1min39sec
30 | 0.740517 | 0.783561 | 0.806966 | 1min9sec
40 | 0.756137 | 0.777656 | 0.806420 | 0min57sec
Inspecting the results from ITEA_regressor and ITExpr_regressor
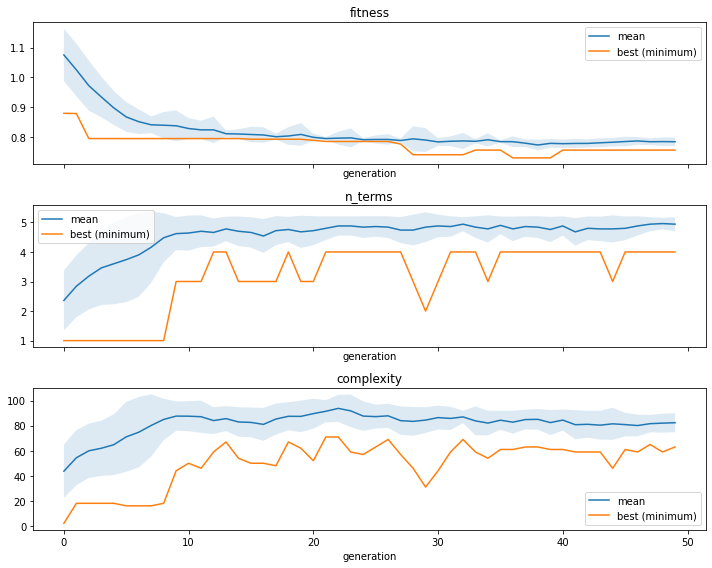
We can see the convergence of the fitness, the number of terms, or tree complexity by using the ITEA_summarizer, an inspector class focused on the ITEA:
[3]:
fig, axs = plt.subplots(3, 1, figsize=(10, 8), sharex=True)
summarizer = ITEA_summarizer(itea=reg).fit(X_train, y_train)
summarizer.plot_convergence(
data=['fitness', 'n_terms', 'complexity'],
ax=axs,
show=False
)
plt.tight_layout()
plt.show()
Now that we have fitted the ITEA, our reg contains the bestsol_ attribute, which is a fitted instance of ITExpr_regressor ready to be used. Let us see the final expression and the execution time.
[4]:
final_itexpr = reg.bestsol_
print('\nFinal expression:\n', final_itexpr.to_str(term_separator=' +\n'))
print(f'\nElapsed time: {reg.exectime_}')
print(f'\nSelected Features: {final_itexpr.selected_features_}')
Final expression:
9.361*log(MedInc^2 * HouseAge * AveRooms * AveBedrms * Population^2 * Latitude * Longitude^2) +
-9.662*log(HouseAge * Population^2 * AveOccup * Longitude^2) +
-8.436*log(MedInc^2 * HouseAge^2 * AveRooms^2 * AveBedrms * Population^2 * Latitude^2) +
-1.954*log(AveRooms) +
8.739*log(HouseAge^2 * AveRooms * Population^2 * AveOccup * Longitude^2) +
-53.078
Elapsed time: 210.53795051574707
Selected Features: ['MedInc' 'HouseAge' 'AveRooms' 'AveBedrms' 'Population' 'AveOccup'
'Latitude' 'Longitude']
[5]:
# just remembering that ITEA and ITExpr implements scikits
# base classes. We can check all parameters with:
print(final_itexpr.get_params)
<bound method BaseEstimator.get_params of ITExpr_regressor(expr=[('log', [2, 1, 1, 1, 2, 0, 1, 2]),
('log', [0, 1, 0, 0, 2, 1, 0, 2]),
('log', [2, 2, 2, 1, 2, 0, 2, 0]),
('log', [0, 0, 1, 0, 0, 0, 0, 0]),
('log', [0, 2, 1, 0, 2, 1, 0, 2])],
labels=array(['MedInc', 'HouseAge', 'AveRooms', 'AveBedrms', 'Population',
'AveOccup', 'Latitude', 'Longitude'], dtype='<U10'),
tfuncs={'cos': <ufunc 'cos'>, 'exp': <ufunc 'exp'>,
'id': <function <lambda> at 0x7f4ab0369680>,
'log': <ufunc 'log'>, 'sin': <ufunc 'sin'>,
'sqrt.abs': <function <lambda> at 0x7f4ab0369f80>})>
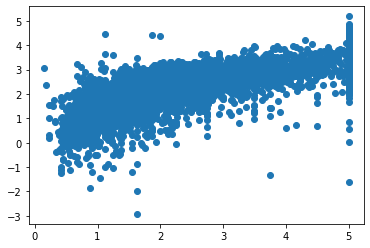
[6]:
fig, axs = plt.subplots()
axs.scatter(y_test, final_itexpr.predict(X_test))
plt.show()
We can use the ITExpr_inspector to see information for each term.
[7]:
display(pd.DataFrame(
ITExpr_inspector(
itexpr=final_itexpr, tfuncs=tfuncs
).fit(X_train, y_train).terms_analysis()
))
| coef | func | strengths | coef\nstderr. | mean pairwise\ndisentanglement | mean mutual\ninformation | prediction\nvar. | |
|---|---|---|---|---|---|---|---|
| 0 | 9.361 | log | [2, 1, 1, 1, 2, 0, 1, 2] | 0.22 | 0.535 | 0.603 | 268.140 |
| 1 | -9.662 | log | [0, 1, 0, 0, 2, 1, 0, 2] | 0.221 | 0.494 | 0.527 | 221.432 |
| 2 | -8.436 | log | [2, 2, 2, 1, 2, 0, 2, 0] | 0.222 | 0.544 | 0.609 | 263.430 |
| 3 | -1.954 | log | [0, 0, 1, 0, 0, 0, 0, 0] | 0.038 | 0.075 | 0.187 | 0.290 |
| 4 | 8.739 | log | [0, 2, 1, 0, 2, 1, 0, 2] | 0.222 | 0.520 | 0.553 | 214.239 |
| 5 | -53.078 | intercept | --- | 1.325 | 0.000 | 0.000 | 0.000 |
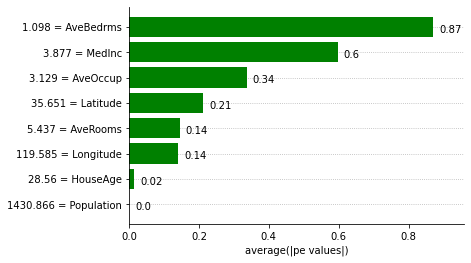
Explaining the IT_regressor expression using Partial Effects
We can obtain feature importances using the Partial Effects and the ITExpr_explainer.
[8]:
explainer = ITExpr_explainer(
itexpr=final_itexpr, tfuncs=tfuncs, tfuncs_dx=tfuncs_dx).fit(X, y)
explainer.plot_feature_importances(
X=X_train,
importance_method='pe',
grouping_threshold=0.0,
barh_kw={'color':'green'}
)
The Partial Effects at the Means can help understand how the contribution of each variable changes according to its values when their covariables are fixed at the means.
[9]:
fig, axs = plt.subplots(2, 4, figsize=(10, 5))
explainer.plot_partial_effects_at_means(
X=X_test,
features=range(8),
ax=axs,
num_points=100,
share_y=False,
show_err=True,
show=False
)
plt.tight_layout()
plt.show()
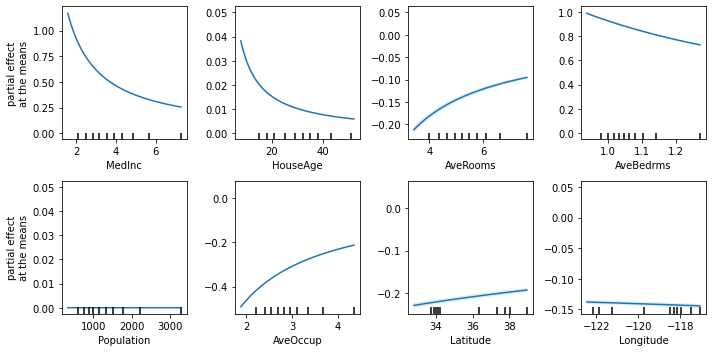
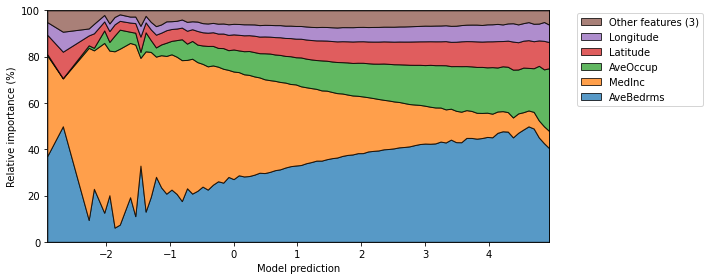
Finally, we can also plot the mean relative importances of each feature by calculating the average Partial Effect for each interval when the output is discretized.
[10]:
fig, ax = plt.subplots(1, 1, figsize=(10, 4))
explainer.plot_normalized_partial_effects(
grouping_threshold=0.1, show=False,
num_points=100, ax=ax
)
plt.tight_layout()